
Getting Started with MacVector: An overview of primer design workflows in MacVector. The alignments can be output in a variety of formats or used to generate phylogenetic reconstructions using a number of built-in algorithms. Melissa Caimano on HOW DO I video guides to common molecular biology workflows MacVector uses the ClustalW algorithm to automatically align any number of sequences and provides a sophisticated editor that lets you fine tune the alignments. admin on HOW DO I video guides to common molecular biology workflows. mariam abdelmalak on Major release details – Summary. Brian on Designing primers and documenting In-Fusion Cloning with MacVector. Chris on Designing primers and documenting In-Fusion Cloning with MacVector.  MacVectorTip: displaying CRISPR PAM Sites on a sequence. The alignment of an address depends on the chosen power of 2. That address is said to be aligned to 4n+3, where 4 indicates the chosen power of 2. for general sequence analysis with phylogeny and alignment programs. For example, the address 0x0001103F modulo 4 is 3. Several equations are included to convert dissimilarity into evolutionary distance. MacVectorTip: Sign up for an NCBI API key to speed up BLAST results Alignment is a property of a memory address, expressed as the numeric address modulo a power of 2. MacVectorTip: Designing Primers for Gibson Assembly. MacVectorTip: Simulating mixed plasmid populations in agarose gels.
MacVectorTip: displaying CRISPR PAM Sites on a sequence. The alignment of an address depends on the chosen power of 2. That address is said to be aligned to 4n+3, where 4 indicates the chosen power of 2. for general sequence analysis with phylogeny and alignment programs. For example, the address 0x0001103F modulo 4 is 3. Several equations are included to convert dissimilarity into evolutionary distance. MacVectorTip: Sign up for an NCBI API key to speed up BLAST results Alignment is a property of a memory address, expressed as the numeric address modulo a power of 2. MacVectorTip: Designing Primers for Gibson Assembly. MacVectorTip: Simulating mixed plasmid populations in agarose gels. Make alignments macvector how to#
MacVectorTip: How to find Restriction Enzymes that only cut outside of a specific region. When you switch to the Picture tab, you will see colored outlines around the shared domains. In the Editor tab a new line will appear above each sequence displaying the extent and color of visible features. In the EDITOR tab using the toolbar button turn on the feature display MODE to SHOW FEATURES. Now run the alignment by clicking ALIGN. 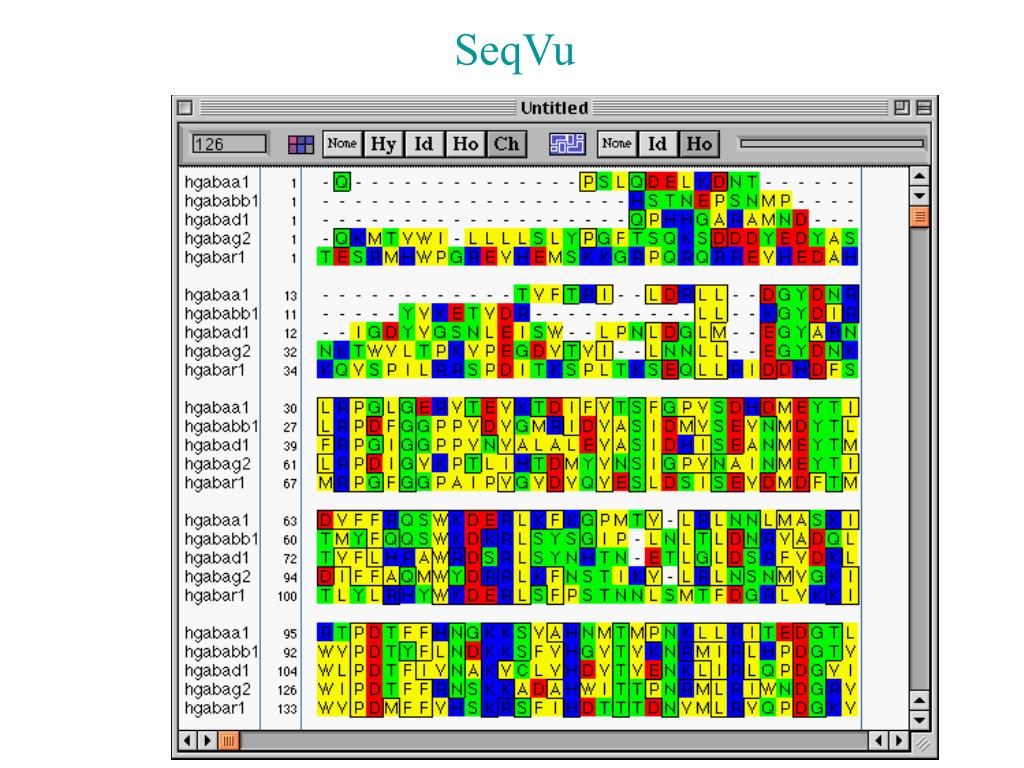
Profile searches of databases, revealing protein motifs (RL) 9.
Click OPTIONS (bottom left hand corner) and choose OPEN MULTIPLE SEQUENCE FILE – AS MULTIPLE ALIGNMENT Multiple sequence alignments, phylogenetic trees (RL) 4, 6 Mar 11 Proteins II. Use FILE | OPEN and select multiple protein sequences. Ensure your protein sequences are annotated and that the domains of interest are visible (you can use DATABASE | INTERPROSCAN to quickly scan and annotate domains to your proteins). Note that alignments created using versions of MacVector before 17.5 will not have this information and will need to be recreated. 
You can also create new domains and dynamically show/hide features in alignments. The colors of features from the individual sequence documents are used to outline the domains in the alignment. MacVector’s new multiple alignment file format retains the features/annotations from the sequences that are used to create the alignment. MacVector has a domain-outlining facility for multiple sequence alignments, letting you easily visualize the relationships between features in aligned protein sequences.






 0 kommentar(er)
0 kommentar(er)
